1.1 Keyword-based Search in TPDdb
TPDdb provides a user-friendly interface with several powerful search options to explore the database. You can perform direct keyword searches for specific molecules or proteins, or use a guided, two-step navigation to browse by biological categories.
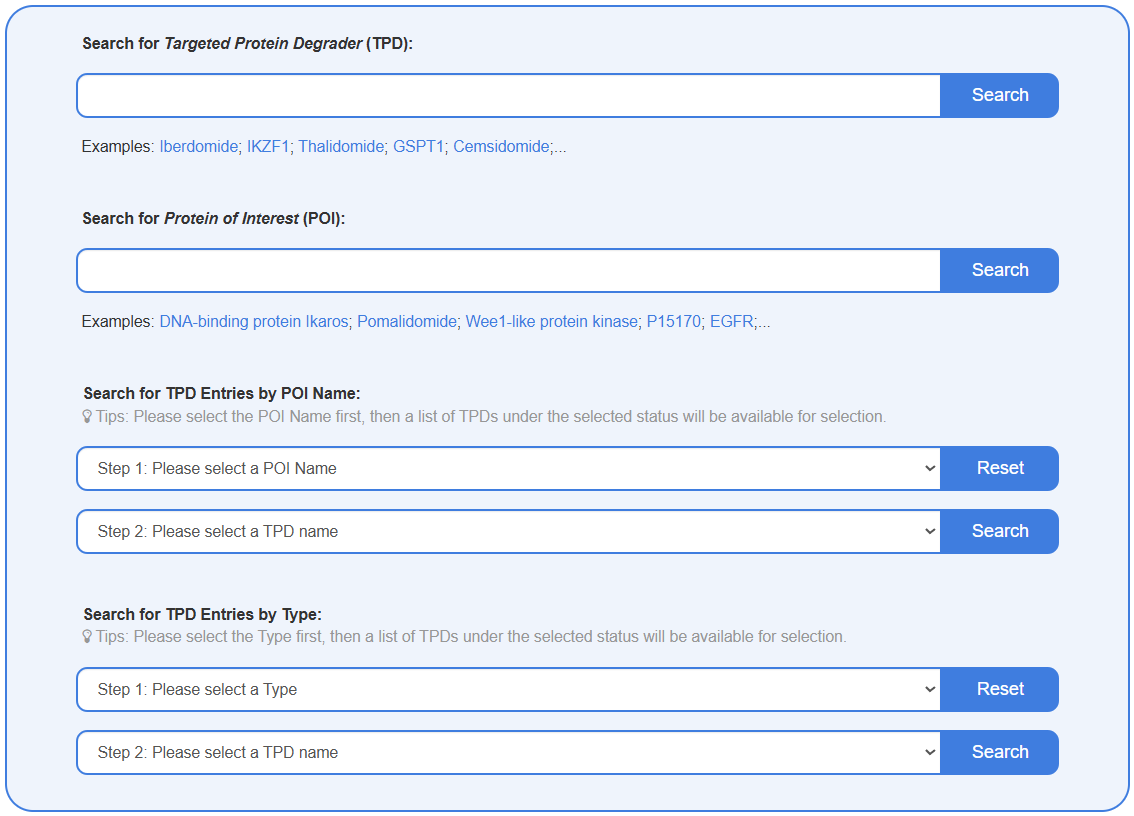
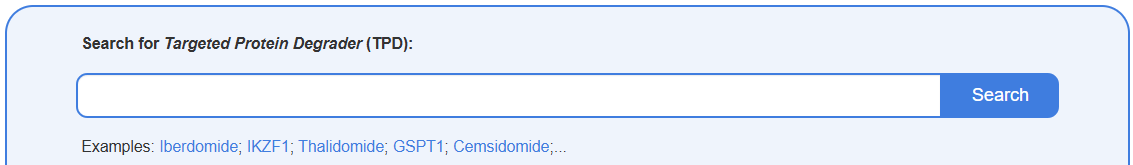
Search for Targeted Protein Degrader (TPD)
This search box allows you to find a specific TPD by entering its name or known synonyms including TPD name, POI ID/name, Ligase ID/name. Upon submission, the results page will display a list of all matching TPDs, showing key information such as TPD Name, TPD Type, and Molecular Formula.
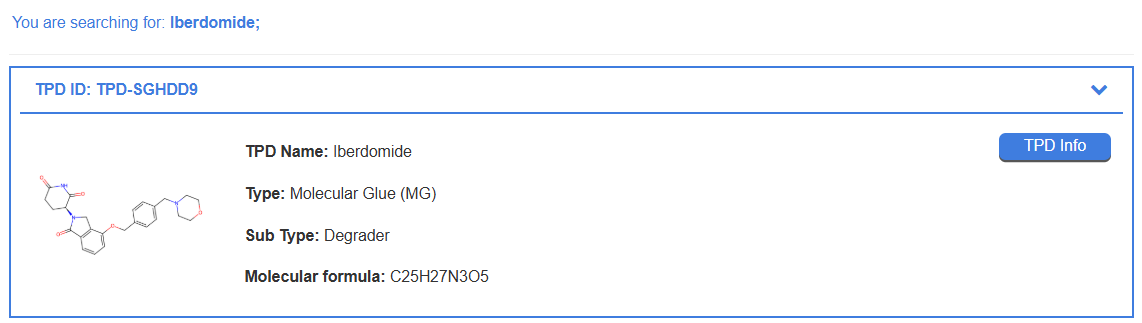
For example, to find detailed information on Iberdomide, simply type "Iberdomide" into the search field. The search result will provide a summary of the molecule. Clicking the “TPD Info” button will direct you to a detailed page with comprehensive data for Iberdomide.
Search for Protein of Interest (POI)
This search box allows you to find information on specific target proteins (POIs) or E3 ligases. You can search by the TPD name, POI ID/name, Ligase ID/name. The results page will display profiles of all proteins matching the search term, including the POI/Ligase Name, Gene Name, and UniProt ID.
For example, to retrieve information about the "DNA-binding protein Ikaros," enter its name into this field. The search results will show its protein name, gene name, and UniProt ID. You can then click the "POI Info" or "Ligase Info" button to access the detailed information page for that specific protein.

Search for TPD Entries by POI Name
This feature enables you to explore TPDs based on their biological target. It uses a two-step process: Step 1: Select a specific POI Name from the comprehensive dropdown list. Step 2: The second menu will then automatically populate with all TPDs in the database known to target that selected POI. Select a TPD name from this list and click search.
This approach is ideal for researchers investigating all known degraders for a particular protein of interest. For instance, to find the TPD named HJM-561, you can first select its target, "ALK tyrosine kinase receptor (ALK)," in Step 1, and then choose "HJM-561" from the list in Step 2.
Search for TPD Entries by Type
This function allows you to browse TPDs based on their modality or classification (e.g., Molecular Glue, PROTAC). This is also a two-step process: Step 1: Select a TPD Type from the dropdown menu, such as "PROteolysis-TArgeting Chimera (PROTAC)". Step 2: The second dropdown menu will then display all TPDs belonging to that category. Select the specific TPD name you are interested in.
This method is particularly useful for exploring the landscape of a specific degradation technology. For example, to find HJM-561, you can select "PROteolysis-TArgeting Chimera (PROTAC)" in Step 1 and then select "HJM-561" in Step 2.
1.2 Structure-based Search in TPDdb
TPDdb has a structure-based search engine to identify TPDs with structural similarity to a query molecule. The search can be performed using three input methods for the query structure. Input a structure by JMSE editor: A chemical structure can be drawn directly using the integrated JSME molecular editor. Upload chemical structure in SDF format: A standard chemical structure file, such as in SDF format, can be uploaded. Input a structure by SMILES: A SMILES string for the molecule of interest can be pasted into the text box.
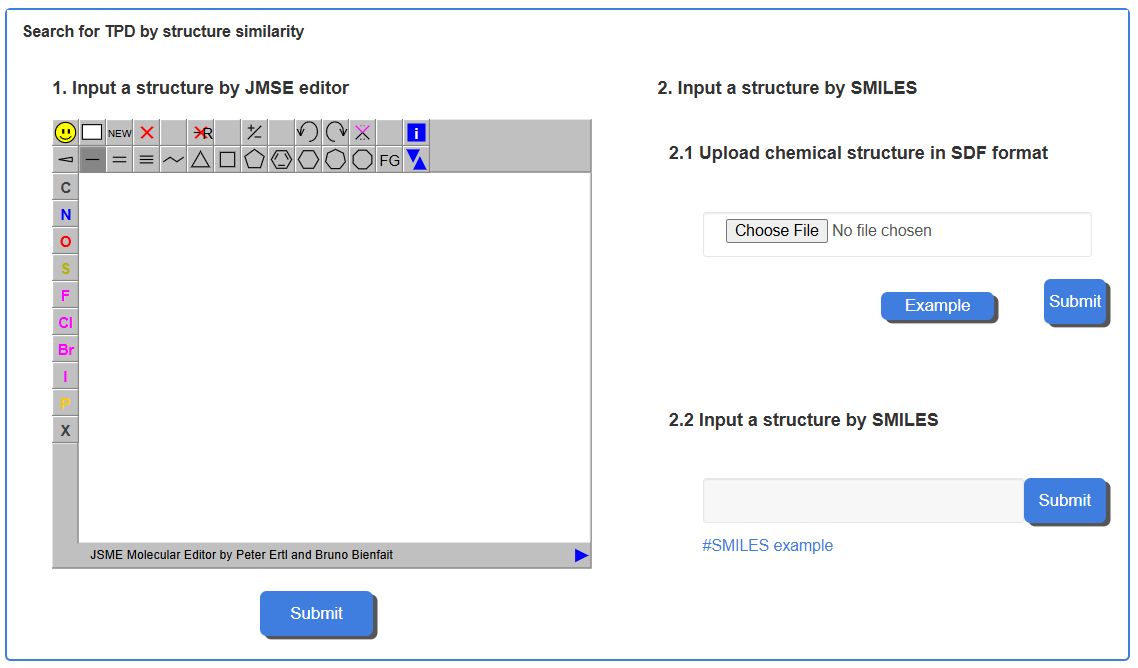
For example, by querying the SMILES, the query molecule is converted into a chemical fingerprint, and its similarity to all molecules in the database is calculated using the Tanimoto coefficient. The search results are presented as a list of molecules ranked by their similarity scores. The results page lists TPDs from the database, and each row includes the TPD ID, a TPD Info button linking to the detailed molecule page, the TPD's Name, its calculated Tanimoto Similarity score, and the Type of Similarity (e.g., High-similarity).
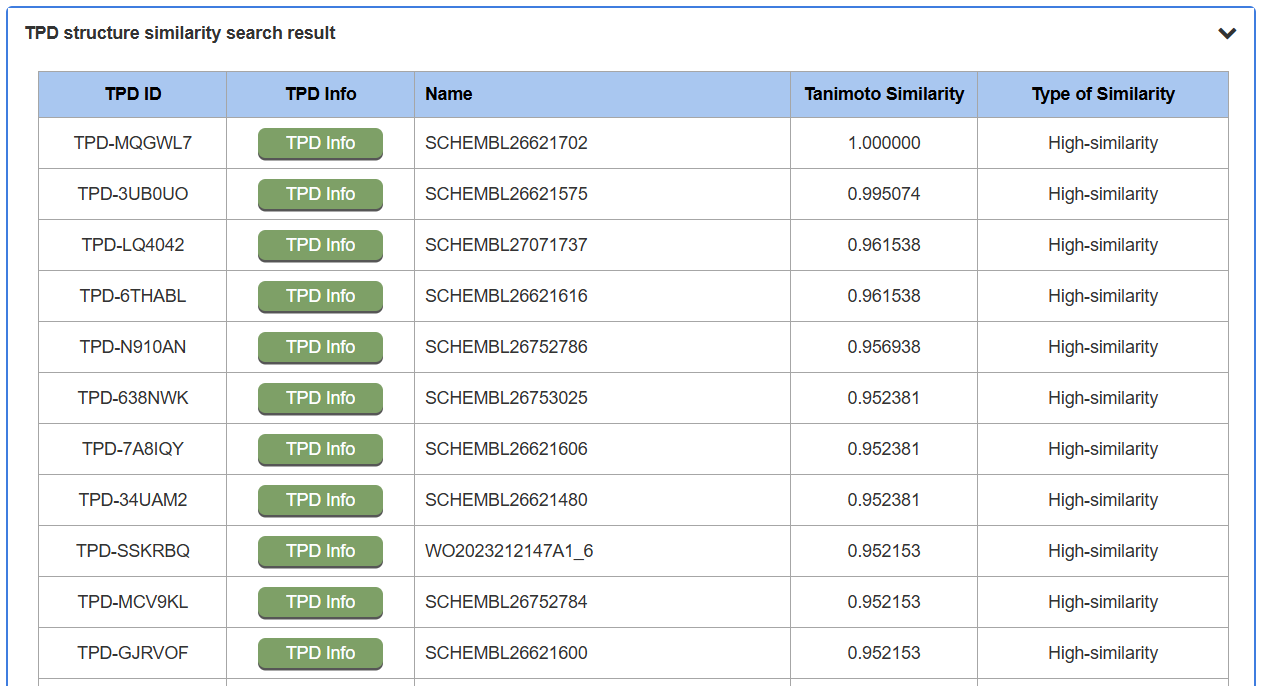
2.1 Detailed TPD Information Page
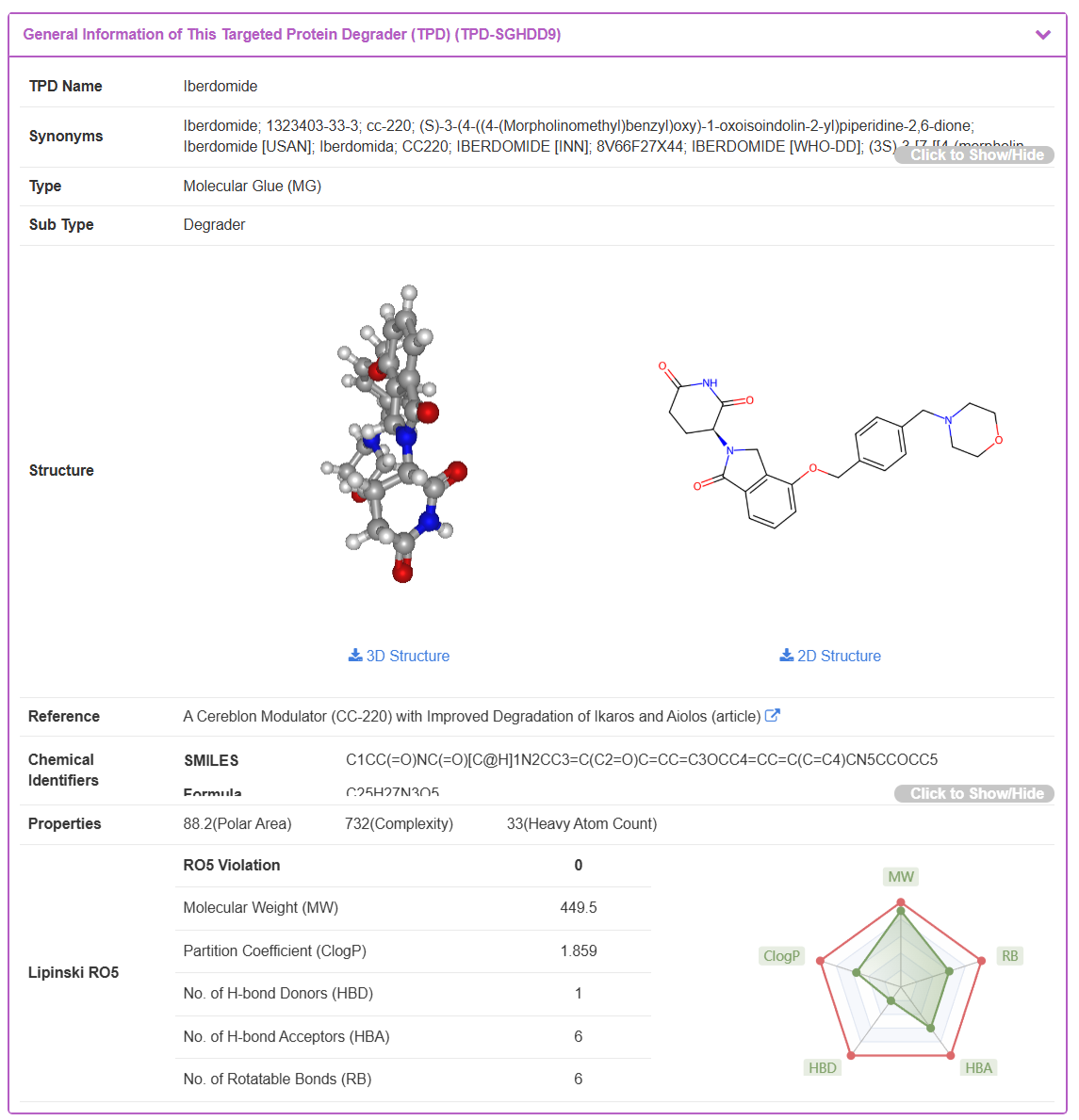
By clicking the “TPD Info” button from any search result, the user is directed to the detailed information page for the selected TPD. Using Iberdomide as an example, this page is organized into several sections providing comprehensive data.
The “General Information of This Targeted Protein Degrader (TPD)” section displays the basic profile of Iberdomide. This includes its TPD Name, Synonyms, TPD Type, interactive 2D and 3D Structures, the original Reference source, Chemical Identifiers (e.g., SMILES, Formula), and calculated physicochemical Properties, including a radar plot for Lipinski's Rule of Five.
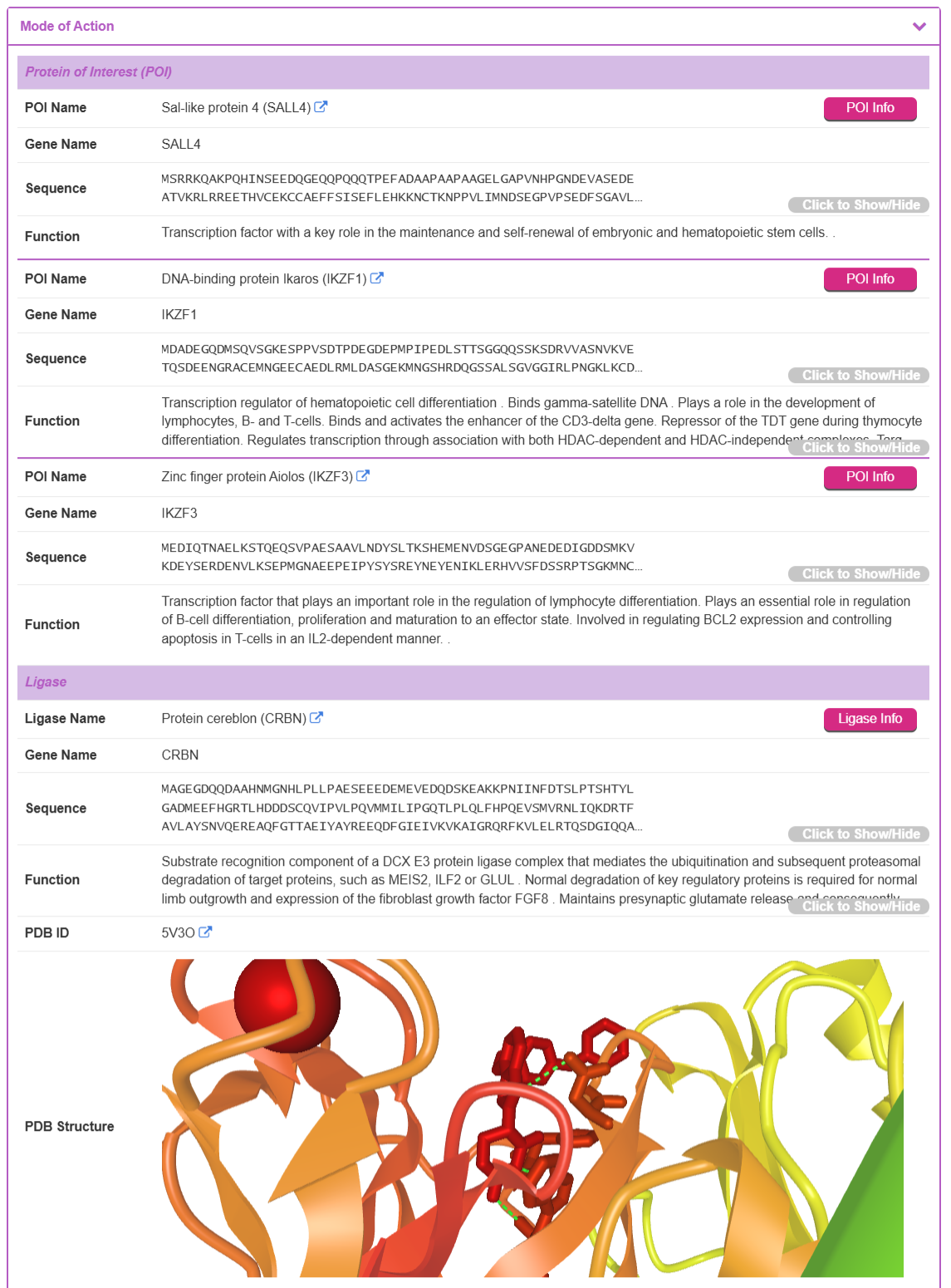
The “Mode of Action” section provides detailed biological and structural context for the TPD. This section is further divided into subsections for POI, Ligase, Binding Preference, and Complex PDB structures.
Protein of Interest (POI) and Ligase: This part lists the target POI(s) and the E3 ligase recruited by the TPD. For each protein, TPDdb provides its POI name, Gene name, Sequence, and Function description. If an experimentally-determined 3D structure is available in the Protein Data Bank (PDB), it is displayed in an interactive viewer. By clicking the “POI Info” or “Ligase Info” button from, the user is directed to the detailed information page for the selected POI or Ligase.
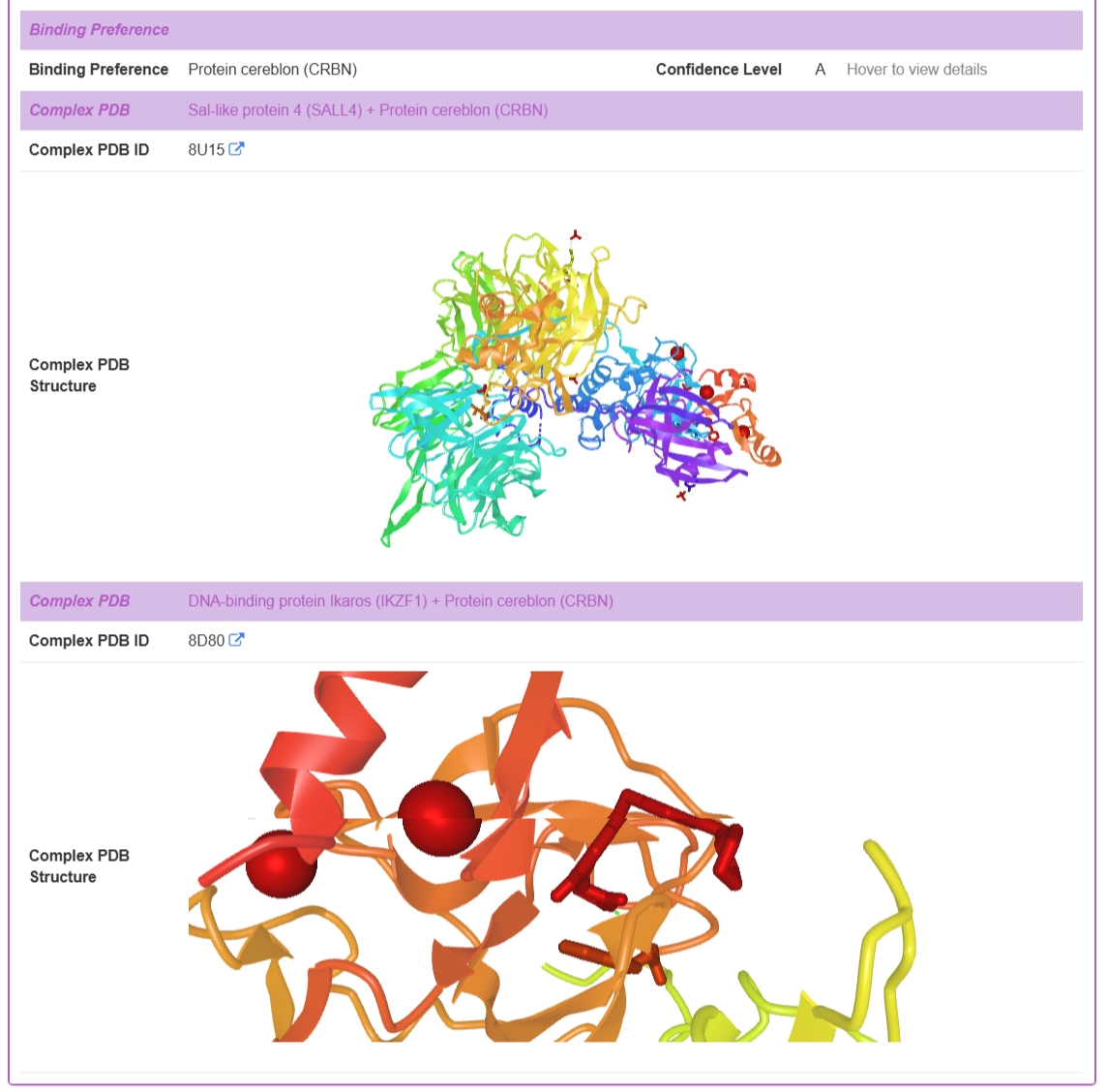
Binding Preference: For Molecular Glues (MGs), TPDdb includes curated binding preference information. Recognizing that the level of evidence for this annotation varies across the literature, a three-tiered confidence scoring system is implemented to provide users with important context about the data's provenance.
Complex PDB Structure: When available, experimentally-determined ternary complex structures (POI-TPD-E3 ligase) are provided. The database displays the Complex PDB ID and an interactive 3D visualization of the complex.
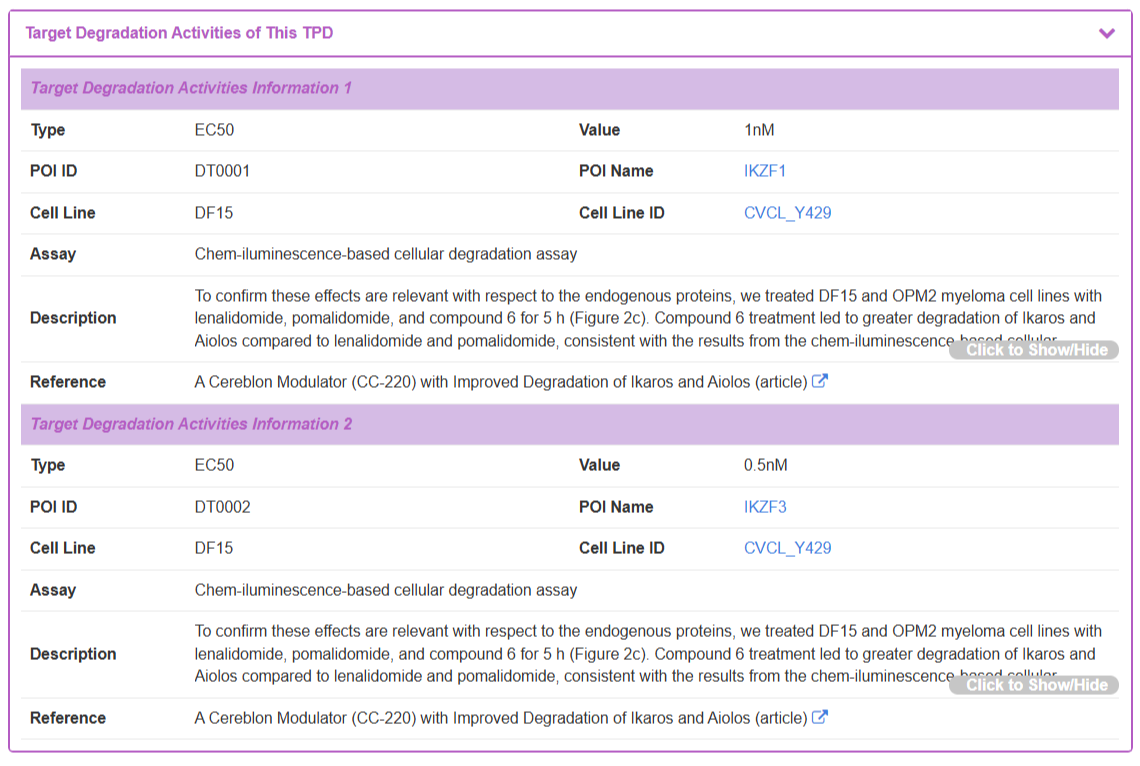
The “Target Degradation Activities” section lists curated activity measurements from biochemical and cellular assays. Each entry provides key experimental context, including the activity Type (e.g., EC50), its measured Value, the specific POI and Cell Line used, the Assay conditions, a Description, and the source Reference.
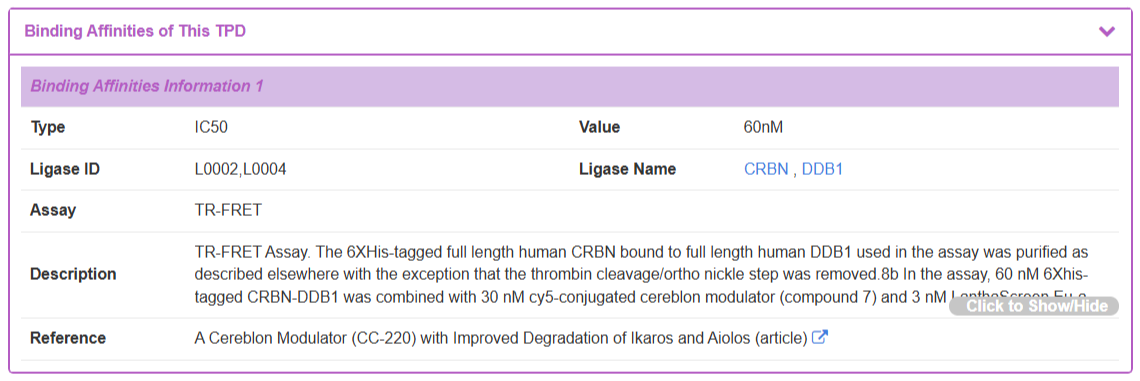
The “Binding Affinities” section, detail the binding activity (e.g., IC50) of the TPD with its target proteins and provide the same level of experimental detail.
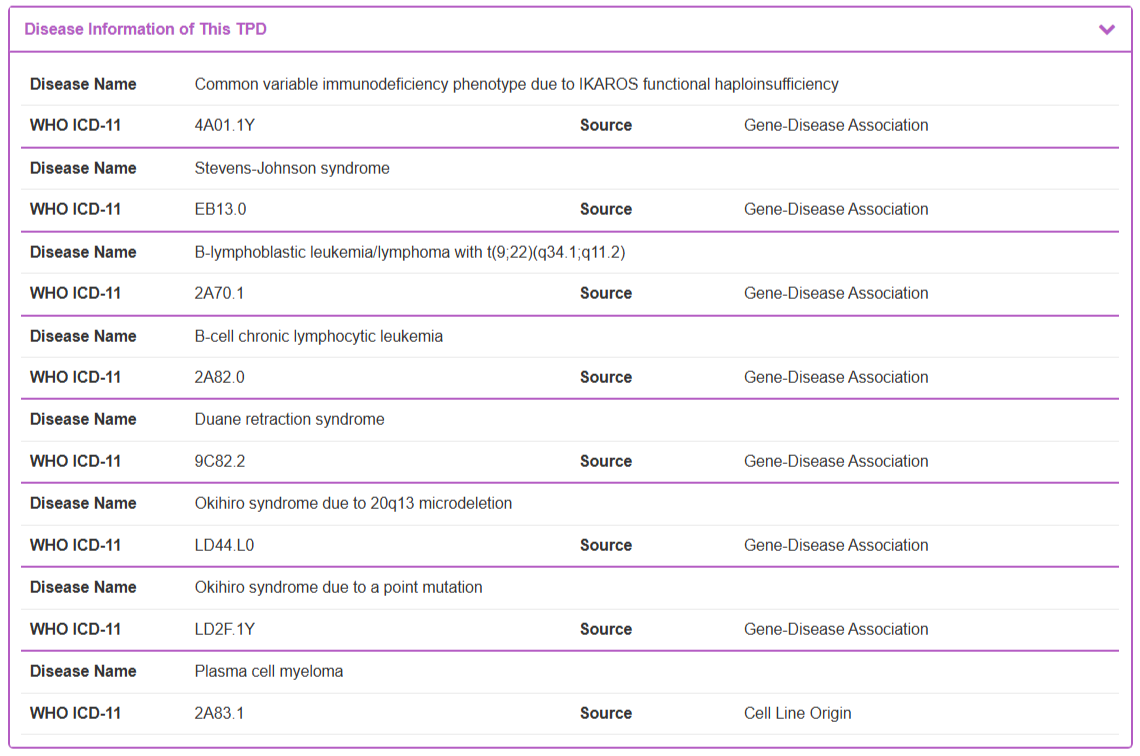
“Disease Information of This TPD” section displays therapeutic context by listing associated diseases. This information is systematically curated using a dual-source strategy. The Source column indicates whether the association is derived from the Cell Line Origin or from established Gene-Disease Associations. All disease names are standardized and presented with their corresponding WHO ICD-11 codes.
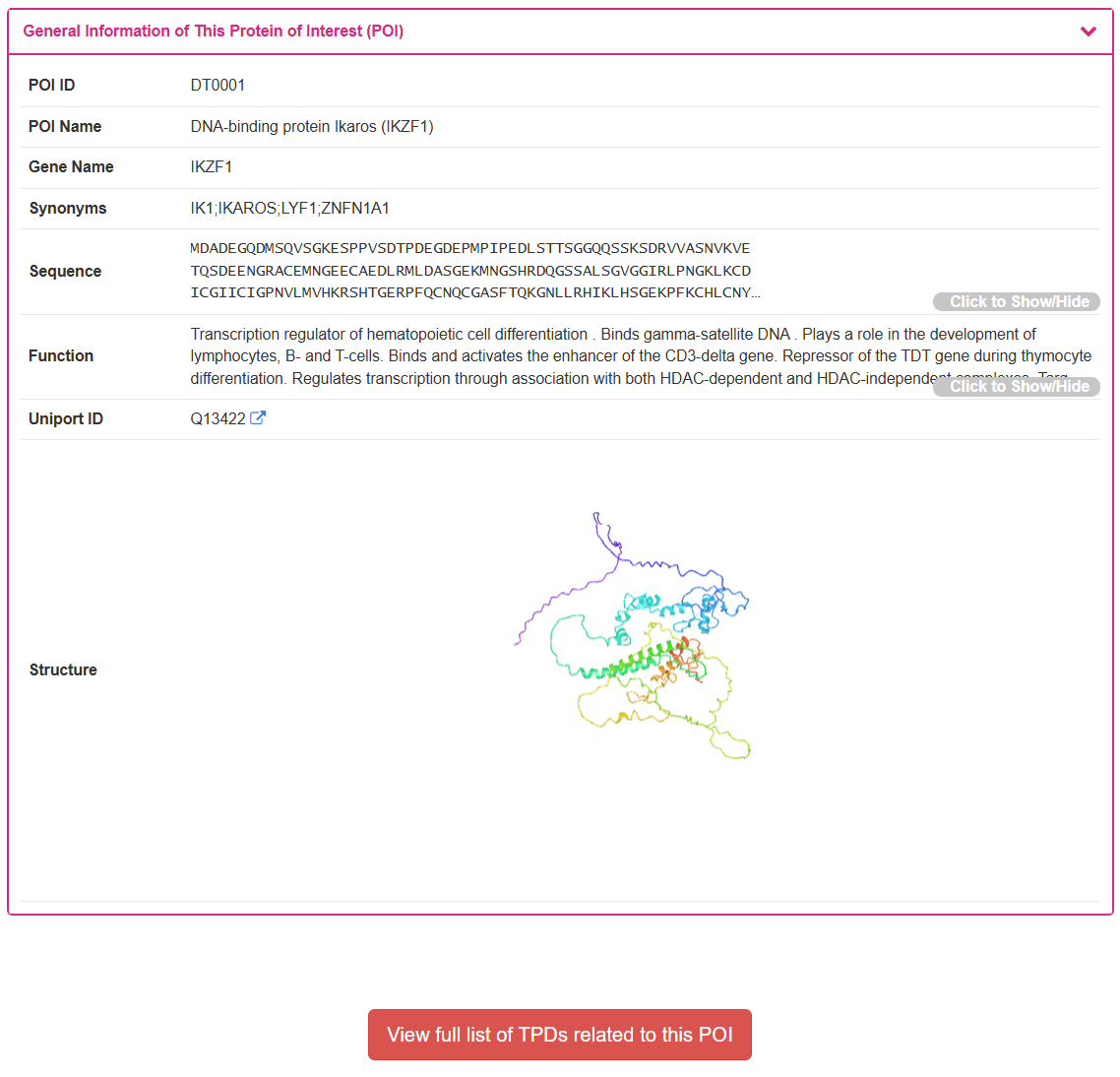
2.2 Detailed POI/Ligase Information Page
By clicking the “POI Info” or “Ligase Info” button from any search result, the user is directed to the detailed information page for the selected POI or Ligase. Using DNA-binding protein Ikaros (IKZF1) as an example, this page displays the general information of this POI including its POI ID, POI Name, Gene Name, and Synonyms. When an experimentally-determined structure is available, it is displayed in an interactive 3D viewer. At the bottom of the page, a button allows the user to “View full list of TPDs related to this POI,” providing a convenient way to explore all degraders targeting this specific protein.